Whole genome multiplication triggers production of evolutionary novel traits
Polyploidisation (or genome multiplication) appears to be absolutely crucial for the evolution of angiosperms. This process is usually accompanied by many fundamental changes, such as an increase in cell size or a change in gene expression, which can subsequently affect plant physiology and morphology. Nevertheless, many questions about polyploidisation remain unresolved, for example, the consequences for survival, coexistence and further reproduction of multiple cytotypes are still a matter of debate.
 Diploid Pilosella rhodopea plant; source: archive of P.Mráz
Diploid Pilosella rhodopea plant; source: archive of P.Mráz
The authors of the study focused on Pilosella rhodopea, a perennial herb from the Balkan Peninsula. This species produces so-called autopolyploids (polyploids within the same species; the opposite is the case for allopolyploids, i.e. polyploids resulting from crosses between two or more species), which are morphologically indistinguishable from their diploid ancestors. In previous studies, the authors have shown that there are three major cytotypes – diploids (those with two sets of chromosomes), triploids (three sets) and tetraploids (four sets of chromosomes), with triploids being the majority cytotype. In this study, the authors collected and examined in detail plants from the Bulgarian mountains. Their main aim was to study the effect of whole genome multiplication on the reproduction of these plants, which could clarify the dominance of triploids and the coexistence of diploids and autopolyploids. "This species is very interesting because it forms the largest primary contact zone between diploids and derived polyploid cytotypes in the world. This 'mixture' is found in virtually every population from which we have analysed a large number of plants for their ploidy level across the Balkan Peninsula," notes co-author of the study Patrik Mráz and further explains, "The cytotypes occur together at the localities, i.e. the autopolyploids are not ecologically segregated from their diploid ancestors and are therefore subject to the same selection pressures. This is a situation that can logically be expected in any primary zone, since autopolyploids should share the same ecological niche as their genetically identical diploid ancestors. Ironically, however, this has not been shown to such an extent in any autopolyploid complex than in Pilosela rhodopea."
 Diploid Pilosella rhodopea plant; source: archive of P.Mráz
Diploid Pilosella rhodopea plant; source: archive of P.Mráz
The results of the study, which was based on a combination of analysis of ploidy of mother plants and their seed progeny, reproductive strategies, embryological observations, and quantification of clonal growth of plants growing in a greenhouse and in one mixed-ploidy population, clearly show that polyploidization disrupts gamete formation, leading to reduced fertility in P. rhodopea autopolyploids. The cause of the low fertility appears to be an unbalanced meiosis, which is manifested by a high production of aneuploid seeds (i.e. seeds in which the cells have an excess or loss of chromosomes compared to the normal haploid set of chromosomes).
A greenhouse experiment revealed that autopolyploids flower less than "normal" diploids, which may be due to the delayed development of autopolyploid plants. The authors also found that, as in similar polyploid complexes, autopolyploid Pilosella rhodopea produce so-called aposporic initials, i.e. cells that can give rise to genetically identical seeds without fertilisation, i.e. apomictically (parthenogenetically). However, in contrast to other model species, this mode of reproduction is not functional in the autopolyploids of P. rhodopea as these cells soon abort. Another interesting fact is that all cytotypes (diploids, triploids and tetraploids) can produce gametes with different numbers of chromosome sets, which further enhances the efficiency of polyploidization in that species. "By the fact that even diploids form unreduced gametes that give rise to de novo autopolyploids, and at the same time there is a frequent gene flow between all cytotypes, their genetic background is the same. This feature, as well as the sharing of an ecological niche between diploids and autopolyploids, provides an excellent opportunity to investigate the direct effects of genome multiplication on phenotype and gene expression, since co-occurring cytotypes differ only in the number of haploid chromosome sets", Patrik Mráz explains, adding that "this model species is truly unique because it allows us to study the early stages of autopolyploid evolution in their natural environment. Thus, it is not necessary to use chemically polyploidized individuals for experiments, because chemical treatment with mitotic inhibitors carries risks of collateral effects on plant growth and physiology."
 Illustration of the results of the greenhouse experiment, source: original study
Illustration of the results of the greenhouse experiment, source: original study
The most interesting result of this study, however, is the finding that autopolyploids produce not only multiple rosettes from axillary buds, but also adventitious rosettes from root buds, which have never been observed in diploid plants. Patrik Mráz explains further:"Immediate formation of these organs as a consequence of polyploidization was also observed in a triploid plant arising from a diploid mother plant, or in triploid plants arising from autopolyploids but this feature in diploid offspring from identical mother autopolyploid plants was absent. When my former Ph.D. student and then postdoc in Fribourg, Switzerland, Barbora Šingliarová and I saw how autopolyploids form dozens of rosettes from root buds and diploids nothing at all, we couldn't believe our eyes. It was like an epiphany. As far as we know, such a qualitative change as a direct consequence of autopolyploidization has not been observed in natural autopolyploid populations before.” Autopolyploids actually compensate for their very low fertility by increased vegetative growth. From an evolutionary point of view, this transition towards increased vegetative growth is crucial, as it allows de novo autolyploids to establish, survive and spread among diploid plants and explains why this primary contact zone is the largest in the world. “Our study also provides an answer to why primary contact zones are a relatively rare phenomenon compared to secondary contact zones. Many autopolyploids are simply not "lucky" enough to have increased clonal reproduction, therefore they cannot take hold in nature, and thus are not recorded by scientists. In short, although Pilosella rhodopea is not a "model Arabidopsis," it provides universal answers to questions about the evolution and ecology of autopolyploids in vascular plants," describes Patrik Mráz.
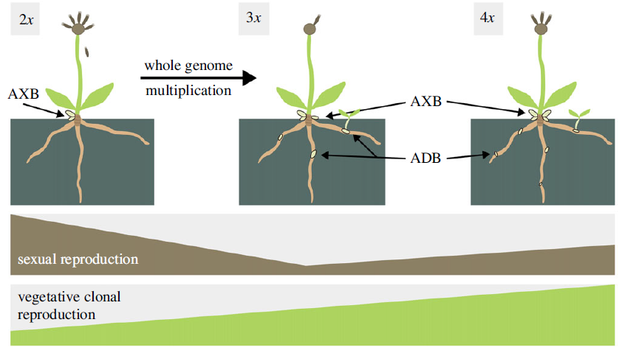 Schematic representation of the differences between diploids and polyploids, source: original study
Schematic representation of the differences between diploids and polyploids, source: original study
"We would like to continue this research. Specifically, we want to focus on the changes that genome multiplication induces at the phytohormone level and the expression of underlying genes, because phytohormones and their interactions regulate plant growth, development and organogenesis. We also want to study the effects of clonal growth, particularly on the age of clones and their spatial structure in natural populations. Preliminary results are very promising, but for a proper and robust study, we would need funding. That's why we have submitted a project proposal on this topic, for the fourth time this year, so hopefully we will finally be lucky", adds Patrik Mráz with a smile.
Šingliarová, B., Hojsgaard, D., Müller-Schärer, H., & Mráz, P. (2023). The novel expression of clonality following whole-genome multiplication compensates for reduced fertility in natural autopolyploids. Proceedings of the Royal Society B, 290(2001), 20230389.
Link to the study here
Document Actions