Peculiarities of the germline-restricted chromosome of songbird
The aptly named germline-restricted chromosome (GRC) was first described 25 years ago in the zebra finch (Taeniopygia guttata). However, recent studies have suggested that it likely occurs in all songbirds, the largest and most diverse bird lineage comprising approximately 50% of all modern bird species. “The GRC is a strange and, in many aspects, contradictory chromosome,“ says Stephen Schlebusch, a postdoc in Radka Reifová’s research group. On one hand, it shows relatively unstable mitotic and meiotic inheritance and can be present in a variable copy number in the male and female germ cells. It also varies greatly in size, being one of the largest chromosomes in the cell in some species, while a tiny microchromosome in other species. This change in size can occur over very short evolutionary times (Figure 1). On the other hand, the GRC has been maintained for more than 47 million years of songbird evolution, suggesting that it is not a mere parasitic chromosome but has some essential function for birds preventing its loss.
 Figure 1. Evolution of the GRC size in five estrildid finches of the genus Lonchura. The GRC is shown in the form of a micronucleus (stained by anti-H3S10P antibody, pink) eliminated from the nucleus of secondary spermatocytes (stained by DAPI, blue). The size of the micronucleus corresponds to the length of the GRC, which can vary from a tiny microchromosome to one of the biggest chromosomes in the cell. The horizontal axis shows millions of years before presence.
Figure 1. Evolution of the GRC size in five estrildid finches of the genus Lonchura. The GRC is shown in the form of a micronucleus (stained by anti-H3S10P antibody, pink) eliminated from the nucleus of secondary spermatocytes (stained by DAPI, blue). The size of the micronucleus corresponds to the length of the GRC, which can vary from a tiny microchromosome to one of the biggest chromosomes in the cell. The horizontal axis shows millions of years before presence.
A previous study from the research team of Alexander Suh, who is also co-author on the Schlebusch et al. paper and is currently based at Leibniz Institute for the Analysis of Biodiversity Change in Germany, showed that the GRC is mostly composed of sequences that were copied on the GRC from regular chromosomes. “This makes attempts to sequence this chromosome challenging, as it is difficult to differentiate the GRC sequences from homologous sequences on regular chromosomes.” says Alexander Suh. For this reason, until recently, only ~ 1% of the zebra finch GRC had been successfully sequenced and assembled.
Schlebusch et al. used a novel strategy and managed to assemble almost the whole GRC in two recently diverged nightingale species, the common nightingale (Luscinia megarhynchos) and the thrush nightingale (L. luscinia). The results were really surprising. Even though the two species diverged less than 2 million years ago and had similarly sized GRCs, they showed dramatic differences in the GRC genetic content (Figure 2). This is striking given that other avian chromosomes are highly collinear, even among distantly related species.
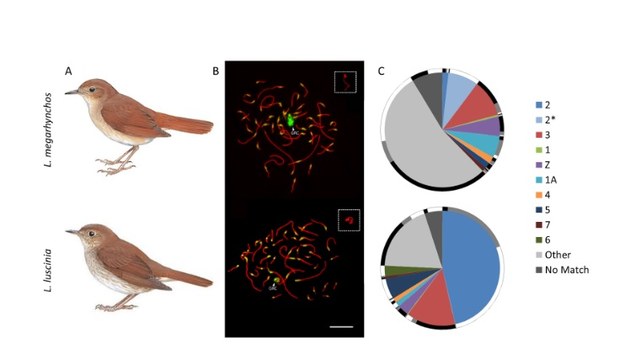 Figure 2. The two nightingale species and their germline-restricted chromosome (GRC). For each species (A) the GRC is visualized in pachytene spermatocytes (B) stained by antibodies labeling the lateral elements of the synaptonemal complex (anti-SYCP3, red) and centromeres (CREST, green). The GRC is marked by arrowheads and shown in higher magnification in the box in the top right corner. (C) The GRC is mostly composed of sequences coming from regular chromosomes (marked in different colors), but contribution of these chromosomes to the GRC dramatically differ in the two species. The homologous sequences between the two GRCs are shown on the outer rim in black, potentially homologous sequences are in grey and species-specific sequences are in white.
Figure 2. The two nightingale species and their germline-restricted chromosome (GRC). For each species (A) the GRC is visualized in pachytene spermatocytes (B) stained by antibodies labeling the lateral elements of the synaptonemal complex (anti-SYCP3, red) and centromeres (CREST, green). The GRC is marked by arrowheads and shown in higher magnification in the box in the top right corner. (C) The GRC is mostly composed of sequences coming from regular chromosomes (marked in different colors), but contribution of these chromosomes to the GRC dramatically differ in the two species. The homologous sequences between the two GRCs are shown on the outer rim in black, potentially homologous sequences are in grey and species-specific sequences are in white.
However, most of the genes on the GRC were found to probably be non-functional, truncated pseudogenes. “It seems that a loss of function is the destiny of most genes that are copied onto the GRC, likely as the result of little selective pressure acting on the GRC due to its presence solely in the germline. You can thus imagine the GRC as a gene graveyard,” explains Stephen Schlebusch.
But not all genes on the GRC are non-functional. From approximately 30 full and possibly functional genes identified on the two nightingale GRCs, about two thirds were present in the entire length only in one of the two species. “This represents a rich substrate for natural selection to act upon in creating reproductive isolation between the species,” says Radka Reifová and further describes the potential evolutionary importance of the GRC: “If similar differences in the GRC genetic content exist among other songbird species, which is likely given the observed rapid GRC size changes, the GRC might significantly speed up the speciation process. This could to some degree explain why songbirds have relatively more species compared to other avian lineages.”
 Figure 3. Common nightingale
Figure 3. Common nightingale
Interestingly, there was only one gene, a paralog of cpeb1 (cytoplasmic polyadenylation element binding protein 1), which was present on the GRC in both nightingale species, showed no copy number variation between individuals and species and had a complete coding region present in all examined individuals. This gene was also identified on the zebra finch GRC. Schlebusch et al. estimated that it was acquired by the GRC early in songbird evolution. This makes this gene one of the oldest genes identified on the GRC and a good candidate for the functional indispensability of the GRC in songbirds. Cpeb1 codes for a mRNA-binding protein that regulates gene expression during oocyte maturation and early embryonic development. At these stages transcription is generally silenced and protein synthesis largely depends on regulation of the translation of stored transcripts, which is controlled by cpeb1. “Although the function of the cpeb1 paralog on the songbird GRC is unknown, we can speculate that it could have specialized in the oocyte-specific function, while the cpeb1 copy on the A chromosomes holds its original functions in somatic cells,” say the authors of the study.
The results described in Schlebusch et al. show that the GRC is a turbulent and rapidly evolving chromosome, which frequently acquires and loses large stretches of sequences duplicated from regular chromosomes, seemingly without consequence for the fitness of the organism. Once on the GRC, most genes appear to lose their function relatively quickly as the restriction to the germline often prevents them from being expressed. In this “gene graveyard”, there is, however, likely a small ancestral region harboring genes that are driving the continued existence of the chromosome in the songbird lineage. “Advances in sequencing technology will enable us to sequence more species and achieve higher quality assemblies, which will allow us to identify this shared region vital to all songbirds” conclude Stephen Schlebusch and Radka Reifová.
 Figure 4. Participants of the GRC workshop held in 2022 at the Faculty of Science, Charles University in Prague. The authors on the Nature Communication paper are marked by an asterisk. From the left: Alexander Suh, Radka Reifová, Tomáš Albrecht, Manon Poignet, Stephen Schlebusch and Francisco Ruiz-Ruano.
Figure 4. Participants of the GRC workshop held in 2022 at the Faculty of Science, Charles University in Prague. The authors on the Nature Communication paper are marked by an asterisk. From the left: Alexander Suh, Radka Reifová, Tomáš Albrecht, Manon Poignet, Stephen Schlebusch and Francisco Ruiz-Ruano.
Schlebusch, S.A., Rídl, J., Poignet, M. et al. Rapid gene content turnover on the germline-restricted chromosome in songbirds. Nat Commun 14, 4579 (2023). https://doi.org/10.1038/s41467-023-40308-8
Document Actions